| DNA Strands | Expected Molecular Weights (no. of bases) |
| 1&2 | 118 |
| 2&3 | 118 |
| 3&4 | 110 |
| 4&5 | 118 |
| 5&6 | 118 |
| 6&1 | 100 |
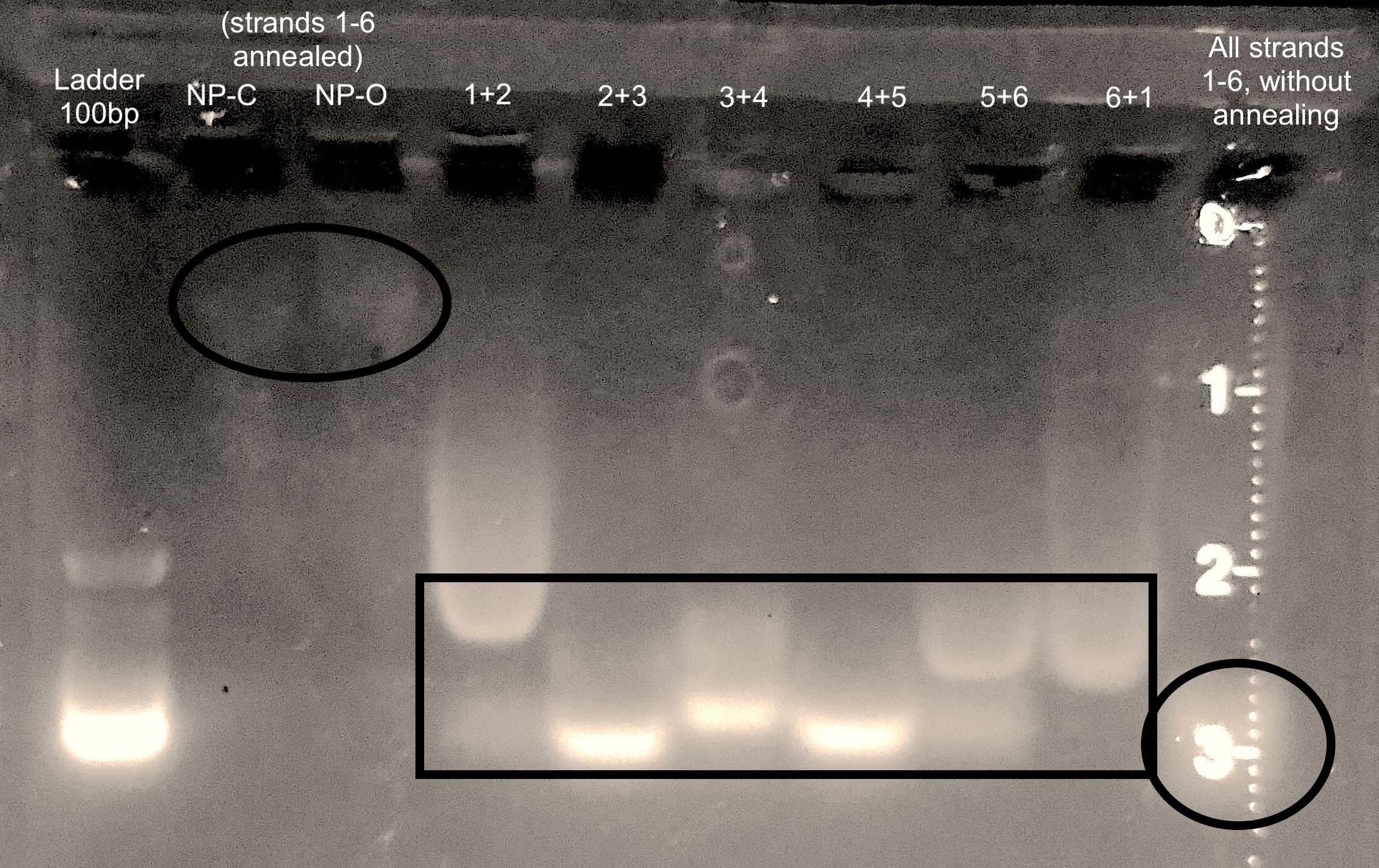 The black box highlights the relative positions of the DNA bands when only two oligonucleotides are annealed together.
The relative differences in these positions correspond to the differences in their molecular weights, as indicated in the table above.
The black circles compare the positions of the DNA bands when all six DNA strands are incubated together, when annealed in a PCR thermocycler (lanes 2 and 3), and withouth annealing in thermocycler (lane 10).
The black box highlights the relative positions of the DNA bands when only two oligonucleotides are annealed together.
The relative differences in these positions correspond to the differences in their molecular weights, as indicated in the table above.
The black circles compare the positions of the DNA bands when all six DNA strands are incubated together, when annealed in a PCR thermocycler (lanes 2 and 3), and withouth annealing in thermocycler (lane 10).
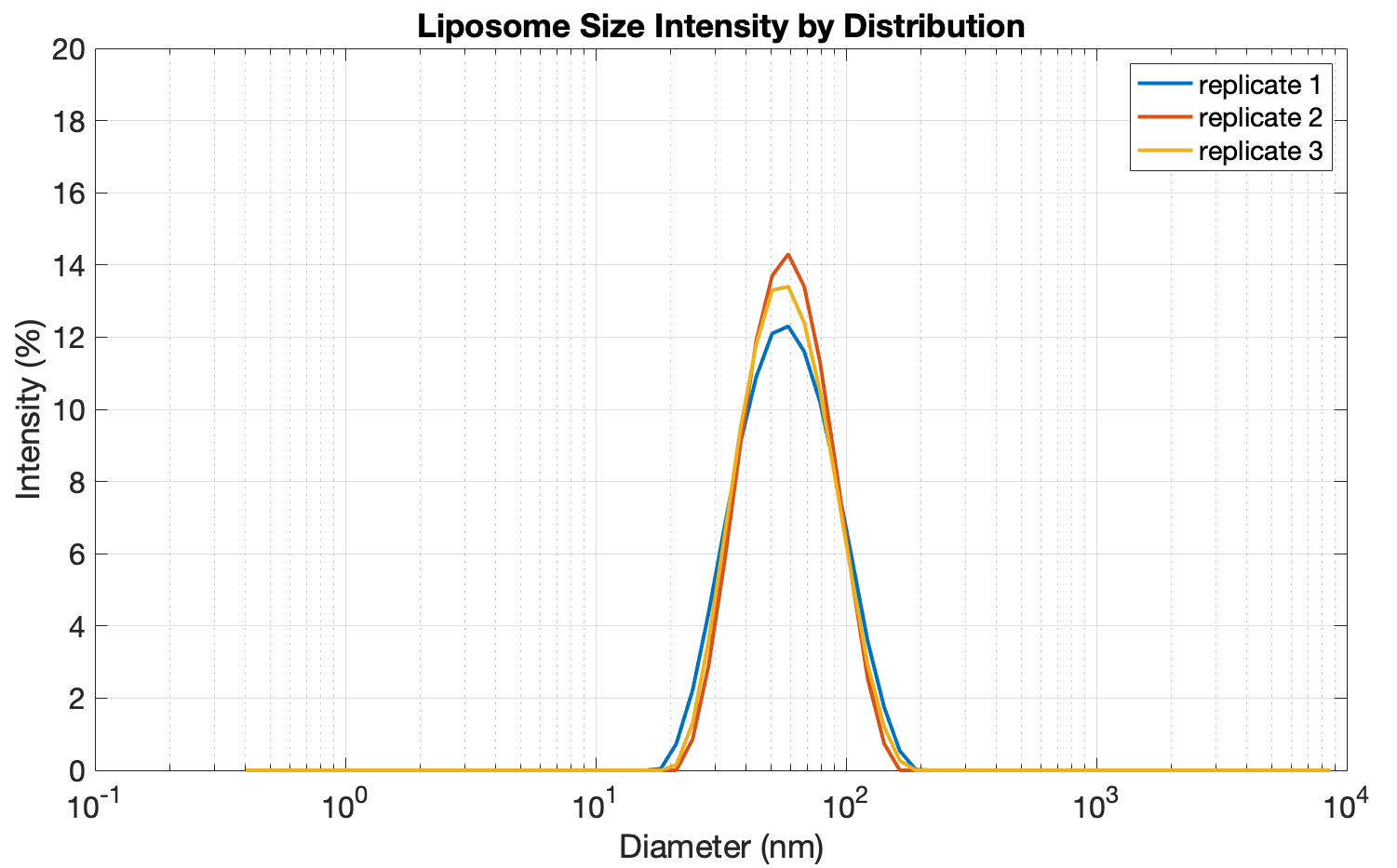
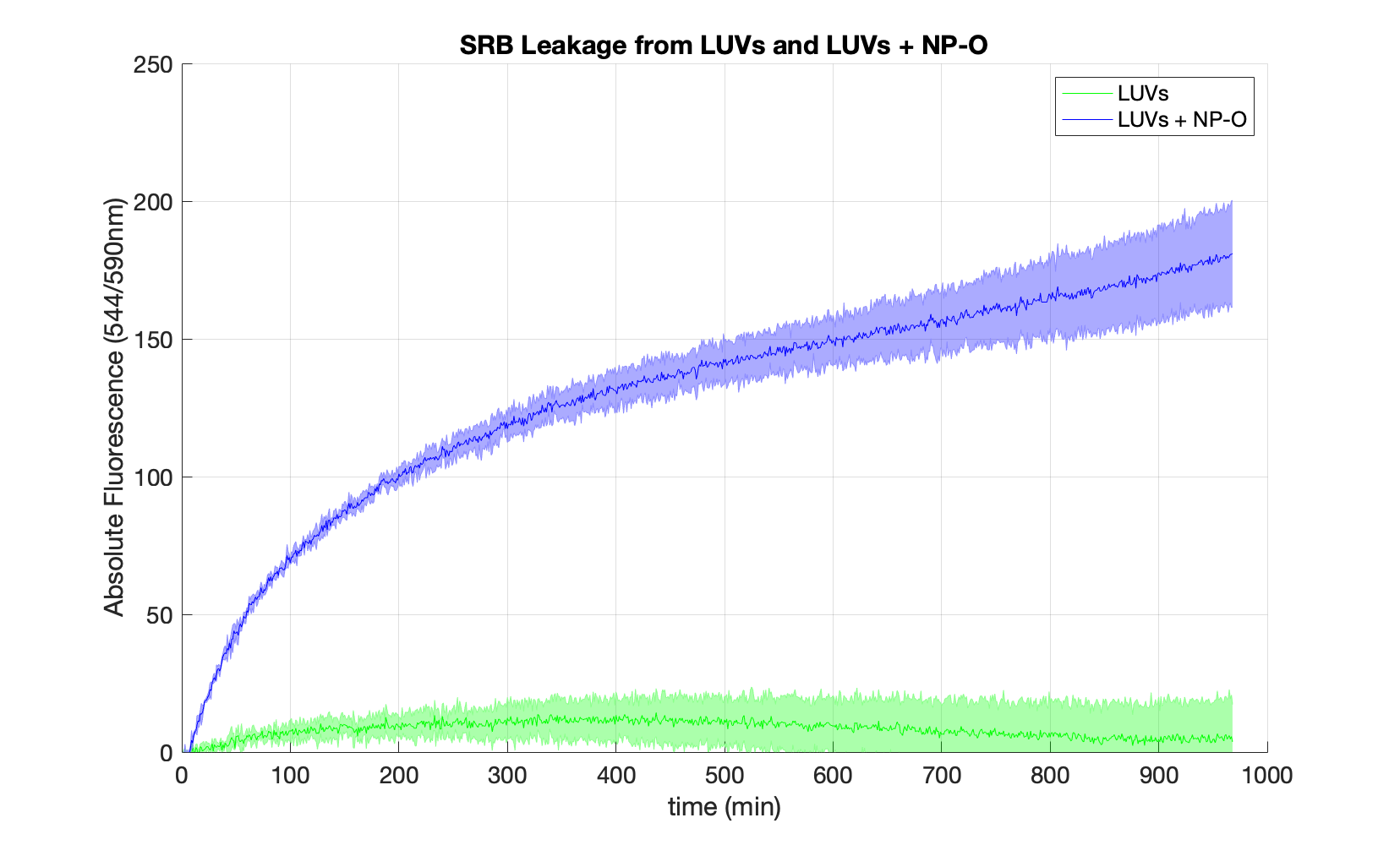 Fluorescence intensity over time of SRB-filled liposomes with (blue line) and without (green line) embedded open nanopores.
Nanopores dissolved in buffer A were added to the phospholipid mixture prior to extrusion. The shaded region corresponds to standard deviation from 3 replicates.
In the beginning the fluorescence levels remained close to zero in both samples due to fluorescence-quenching.
However, when open nanopores are present in the membrane, SRB leaks from the vesicles over time. As it is no longer trapped inside in a high-concentration environment, the fluorescence of the sample increases until reaching a steady state after ca. 600 min. If no nanopores are embedded into the membrane, SRB stays inside the vesicle with minimal leakage (green curve).
Taken together, these data demonstrates that the assembled nanopores can emben into the membrane of LUVs, allowing SRB to leak out.
Fluorescence intensity over time of SRB-filled liposomes with (blue line) and without (green line) embedded open nanopores.
Nanopores dissolved in buffer A were added to the phospholipid mixture prior to extrusion. The shaded region corresponds to standard deviation from 3 replicates.
In the beginning the fluorescence levels remained close to zero in both samples due to fluorescence-quenching.
However, when open nanopores are present in the membrane, SRB leaks from the vesicles over time. As it is no longer trapped inside in a high-concentration environment, the fluorescence of the sample increases until reaching a steady state after ca. 600 min. If no nanopores are embedded into the membrane, SRB stays inside the vesicle with minimal leakage (green curve).
Taken together, these data demonstrates that the assembled nanopores can emben into the membrane of LUVs, allowing SRB to leak out.
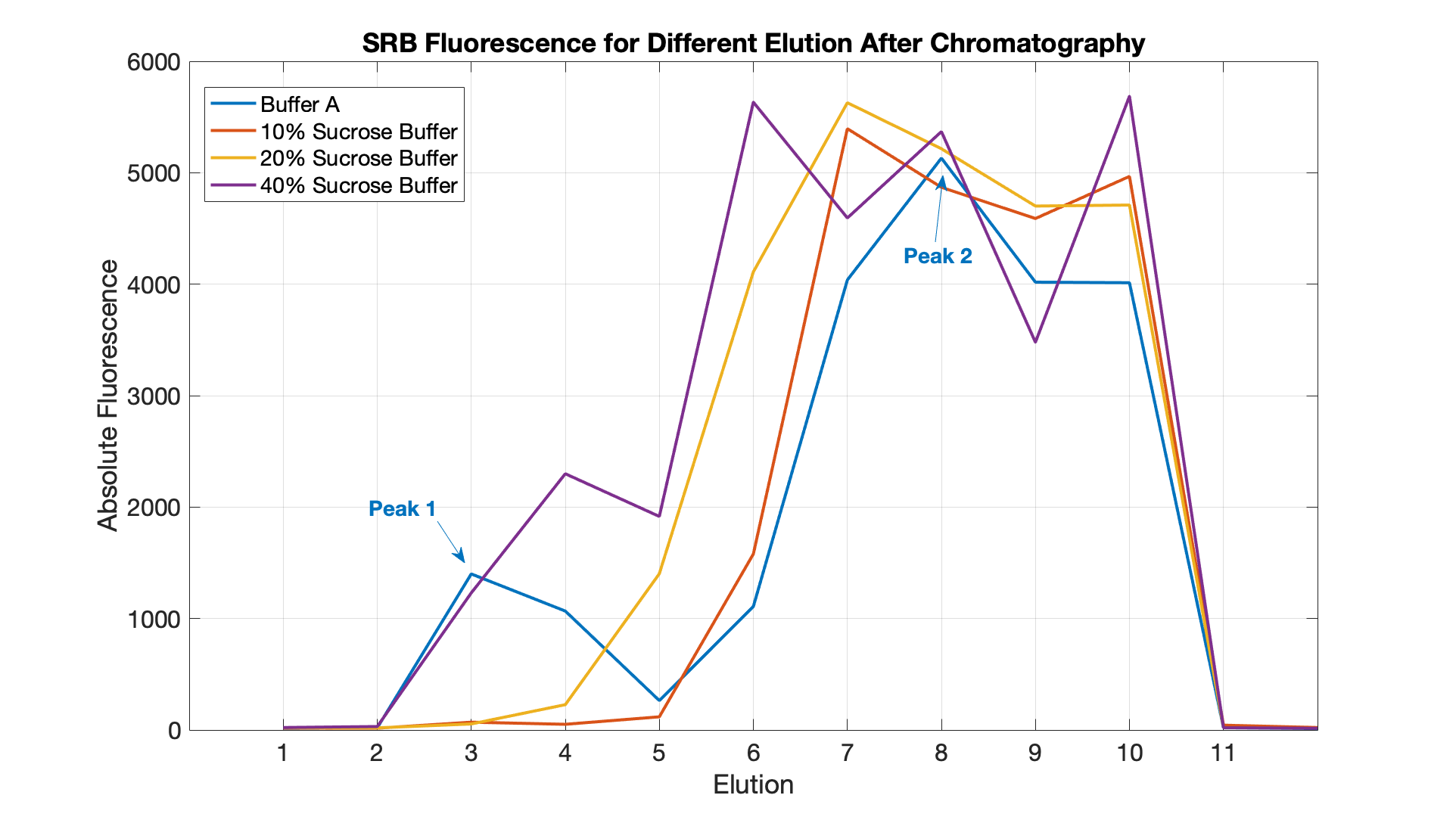
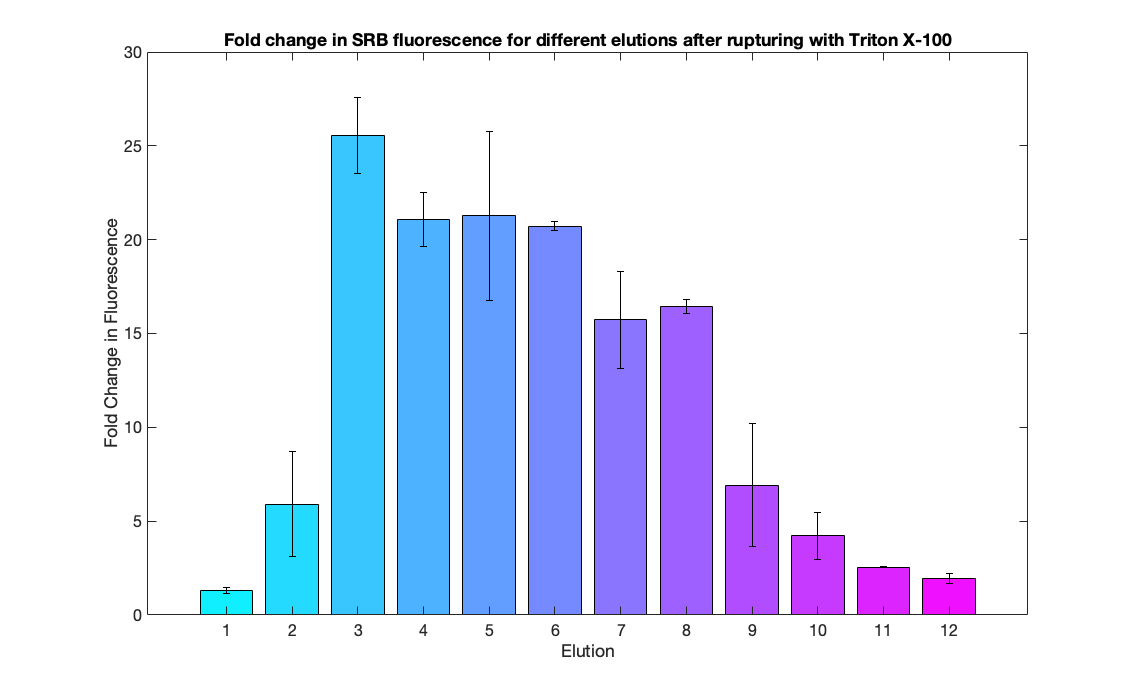
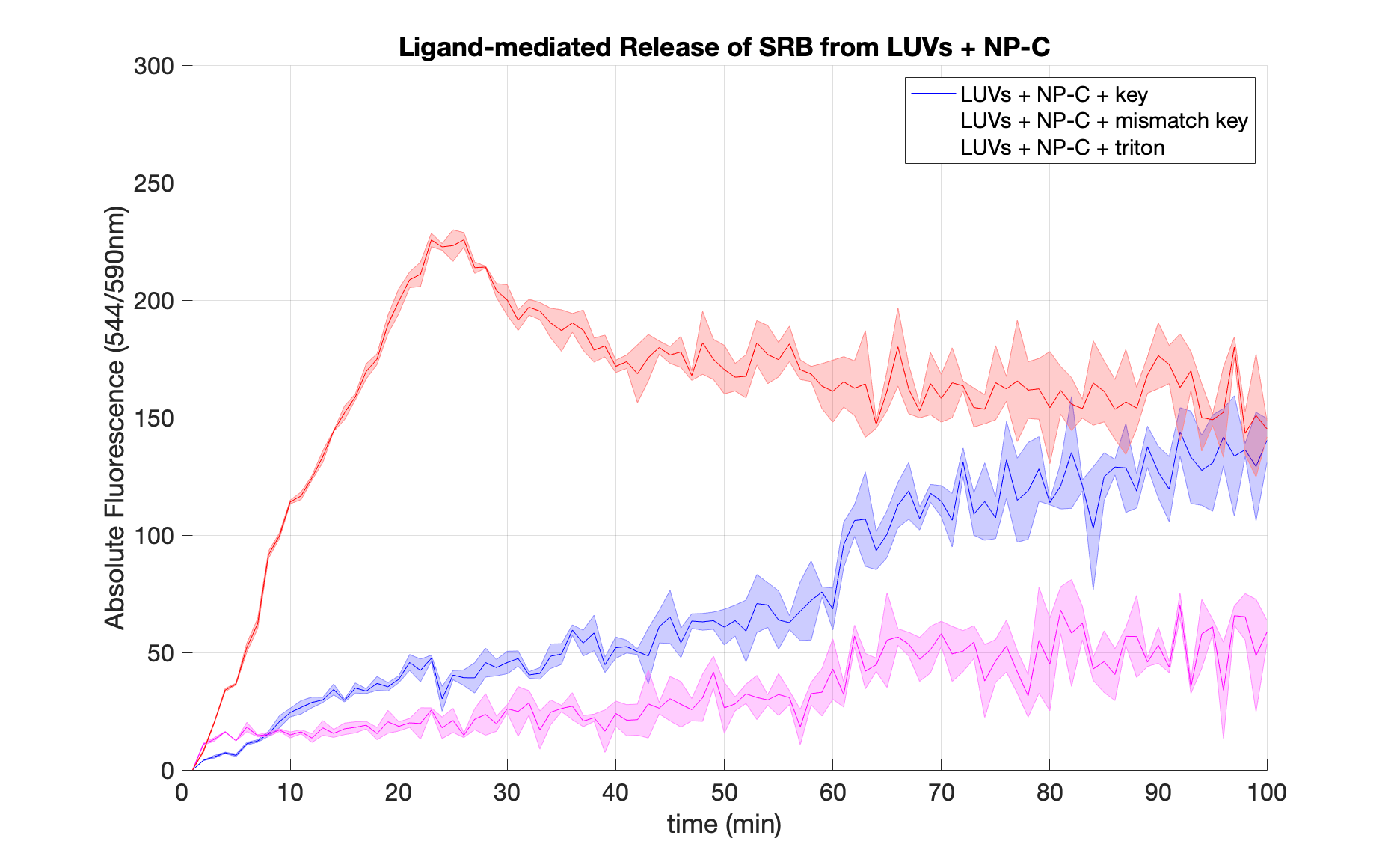 We demonstrated the specificity of ligand-mediated opening of closed nanopores by comparing the effects of adding a key with perfect complementarity to the lock (blue curve) and a key containing mismatches in the sequence (pink curve). To obtain an endpoint value for the fluorescence, we treated nanopore-containing vesicles with 5 ul of 20% Triton X-100 (red curve).
When the exact key is added, SRB is released from liposomes at a higher rate compared to the mismatch key. As a result, after 100 mins the abosulte fluorescence of the sample with the matching key ris approximately 3 times higher than the LUVs-NP-C containing the mismatch key.
The dramatic increase in fluorescence following the addition of Triton X-100 proved the encapsulation of SRB. The drop in fluorescence that follows may be explained by self-quenching at very high SRB concentrations.
We demonstrated the specificity of ligand-mediated opening of closed nanopores by comparing the effects of adding a key with perfect complementarity to the lock (blue curve) and a key containing mismatches in the sequence (pink curve). To obtain an endpoint value for the fluorescence, we treated nanopore-containing vesicles with 5 ul of 20% Triton X-100 (red curve).
When the exact key is added, SRB is released from liposomes at a higher rate compared to the mismatch key. As a result, after 100 mins the abosulte fluorescence of the sample with the matching key ris approximately 3 times higher than the LUVs-NP-C containing the mismatch key.
The dramatic increase in fluorescence following the addition of Triton X-100 proved the encapsulation of SRB. The drop in fluorescence that follows may be explained by self-quenching at very high SRB concentrations.
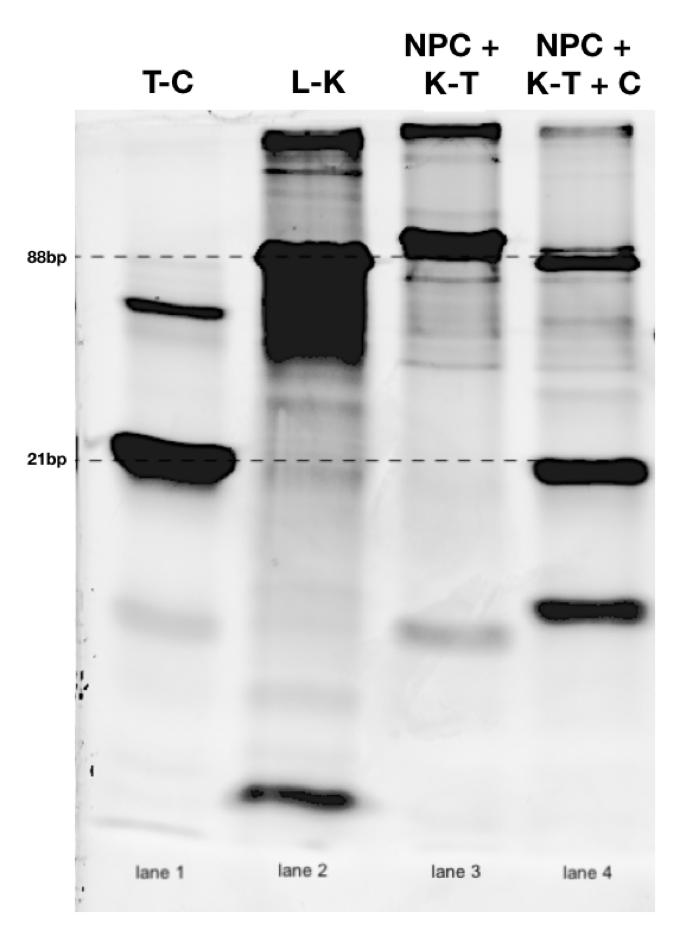 Strand displacement mediated opening of DNA nanopore. A 25% polyacrylamide gel was used. In lane 4, the complement (C) strand would theoretically bind the toehold (T), displacing the key (K). This would allow the key to bind the lock strand. As a result, DNA bands corresponding to the following complexes should be visible: 21bp toehold-complement hybrid (K-T) and 88bp lock-key hybrid (L-K).
Lanes 1-3 are controls. T-C = annealed toehold and complement DNA strands; L-K = annealed lock and key DNA strands; NPC = annealed closed nanopore; K-T = annealed key and toehold DNA strands; C = complement DNA strand. DNA strands were annealed in a PCR thermocycler as described in the (Build) page.
Strand displacement mediated opening of DNA nanopore. A 25% polyacrylamide gel was used. In lane 4, the complement (C) strand would theoretically bind the toehold (T), displacing the key (K). This would allow the key to bind the lock strand. As a result, DNA bands corresponding to the following complexes should be visible: 21bp toehold-complement hybrid (K-T) and 88bp lock-key hybrid (L-K).
Lanes 1-3 are controls. T-C = annealed toehold and complement DNA strands; L-K = annealed lock and key DNA strands; NPC = annealed closed nanopore; K-T = annealed key and toehold DNA strands; C = complement DNA strand. DNA strands were annealed in a PCR thermocycler as described in the (Build) page.
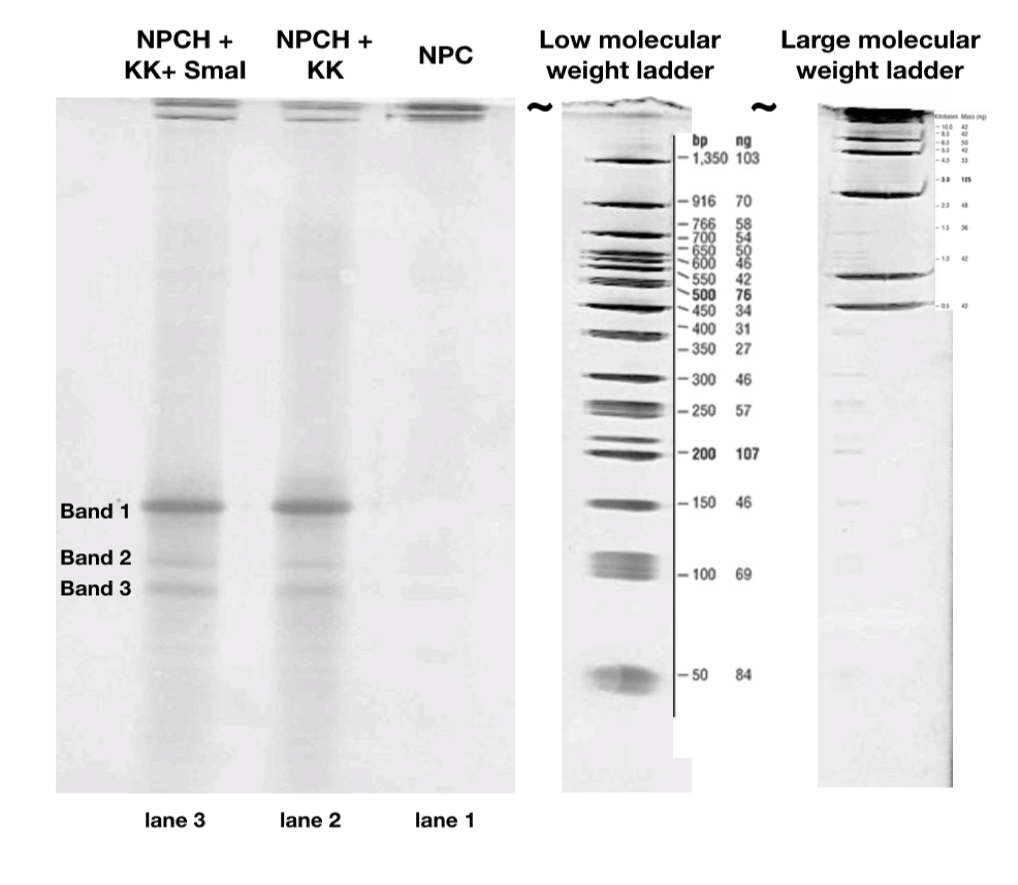 A 15% polyacrylamide gel was used. From right to left: In the control lane 1, no bands are visible except for the darkly staining well, indicating the presence of an intact closed nanopore (NPCH). In control lane 2, NPCH is incubated with a DNA sequence designated Key Kite (KK). In lane 3, the restriction enzyme SmaI is also added. The KK consists of a SmaI recognition site, which can trigger the downstream displacement of DNA strands on the KK to ultimately release the key DNA strand. In lanes 2 and 3, band 1 likely corresponds to a 153bp complex of the 97bp KK and 56bp lock. This indicates promiscuous, undesired KK activity regardless of SmaI presence. Bands 2 and 3 likely correspond to lock-key and complement-toehold complexes. Notably, these bands are stronger in lane 3, when the SmaI is added. This is in spite of the volume of KK used in lane 3 being half that used in lane 2. KK and NPCH were separately annealed in a PCR thermocycler as described in the Build page. NPCH was incubated with KK, or with KK+SmaI for 30 minutes at room temperature prior to running the gel. The ladders shown belong to the same gel.
Our experiments indicate a certain degree of promiscuous activity of the modified DNA sequences, resulting in some undesired lock-key complexes (Fig.8).
Future research would involve further characterisation of the bands corresponding to such suspected promiscuous activity to improve the design of the sequences.
A 15% polyacrylamide gel was used. From right to left: In the control lane 1, no bands are visible except for the darkly staining well, indicating the presence of an intact closed nanopore (NPCH). In control lane 2, NPCH is incubated with a DNA sequence designated Key Kite (KK). In lane 3, the restriction enzyme SmaI is also added. The KK consists of a SmaI recognition site, which can trigger the downstream displacement of DNA strands on the KK to ultimately release the key DNA strand. In lanes 2 and 3, band 1 likely corresponds to a 153bp complex of the 97bp KK and 56bp lock. This indicates promiscuous, undesired KK activity regardless of SmaI presence. Bands 2 and 3 likely correspond to lock-key and complement-toehold complexes. Notably, these bands are stronger in lane 3, when the SmaI is added. This is in spite of the volume of KK used in lane 3 being half that used in lane 2. KK and NPCH were separately annealed in a PCR thermocycler as described in the Build page. NPCH was incubated with KK, or with KK+SmaI for 30 minutes at room temperature prior to running the gel. The ladders shown belong to the same gel.
Our experiments indicate a certain degree of promiscuous activity of the modified DNA sequences, resulting in some undesired lock-key complexes (Fig.8).
Future research would involve further characterisation of the bands corresponding to such suspected promiscuous activity to improve the design of the sequences.