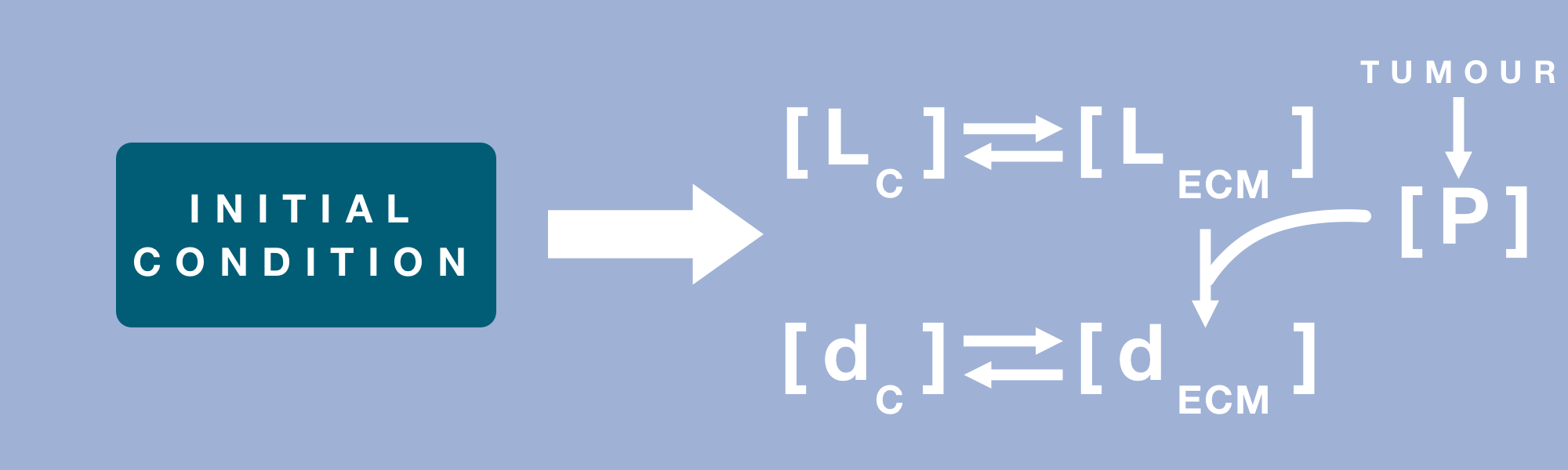
Where the species in the system can be identified using table 1 below.
| Term | Represents |
|---|---|
| dECM | Drug molecule in ECM |
| dC | Drug molecule in capillary |
| LECM | Liposome in ECM |
| LC | Liposome in capillary |
| P | Protease |
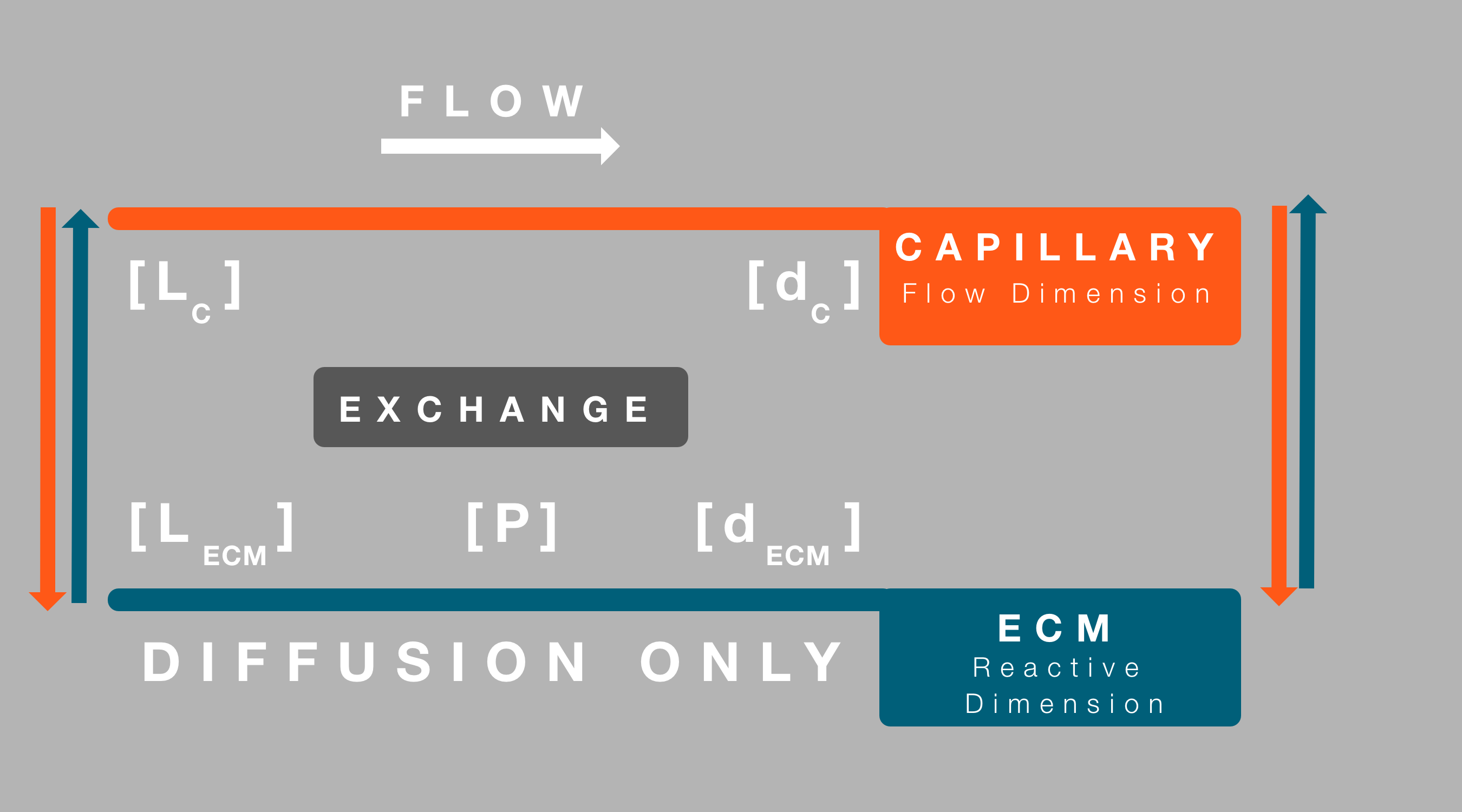
Simplifications included examining the system as a one-dimensional state, where a single capillary traversed space parallel to this dimension, while surrounded by Extra Cellular Matrix (ECM). The flow of species within the system was appropriately simplified based on the following conditions:
| Term | Value (mm2 s-1) | Represents |
|---|---|---|
| Dd-ECM | 1 x 10-4 (Swabb et al. 1974) | Diffusivity of drug molecules in ECM |
| Dd-C | 1 x 10-3 | Diffusivity of drug molecules in capillary |
| D*L-ECM | 1 x 10-5(Murray et al. 1992) | Diffusivity of liposomes in ECM |
| D*L-C | 1 x 10-4 | Diffusivity of liposomes in capillary |
| DP | 1 x 10-4(Hettiaractchi et al. 2018) | Diffusivity of protease in ECM |
* Due to the size of the liposomes produced (diameter = ~150 nm), the order of magnitude for their diffusivity was based upon virus cells of similar size.
Values for the drug in the system are based upon the Cisplatin molecule currently used as a chemotherapy product. Values for diffusivity in mammalian tissues are well documented in literature and used accordingly for this model, however, finding values for diffusivity in blood medium produced less relevant results. As in the capillary region of the model, the order of magnitude of the flow term present dominates the diffusion term (based on scaling analysis of the governing PDE), it was less pertinent to find accurate values for diffusivity in blood. It was therefore assumed that the diffusivity increased 10-fold when in blood, compared to ECM, due to being in fluid medium.
Representing the complex process of exchange between the two regions in the system was critical. To minimise the complexity of modelling these processes, exchange was idealised as a one-step process with a constant rate coefficient describing it. Enabling the exchange processes to be represented in the PDEs concisely. Documented below is the rate coefficients used to describe the one-step exchanges and other key behaviours in the system, again assumed to be constant in space and time.
| Term | Value (s-1) | Represents |
|---|---|---|
| kd-Ex-ECM | 1 x 10-2 * | Rate coefficient for drug molecule exchange into ECM |
| kd-Ex-C | 1 x 10-2 | Rate coefficient for drug molecule exchange into capillary |
| kL-Ex-ECM | 5 x 10-2 | Rate coefficient for liposome exchange into ECM/td> |
| kL-Ex-C | 5 x 10-2 | Rate coefficient for liposome exchange into capillary |
| Deathd | 1.5 x 10-4(Nagai et al., 1996) | Rate coefficient of natural decomposition of drug molecules |
| DeathL | 1 x 10-2 | Rate coefficient of natural decomposition of liposomes |
| DeathP | 1 x 10-3(Craik et al. 2015) | Rate coefficient of natural decomposition of protease |
| kP-Prod | 5 x 10-3 M | Rate coefficient for the constant production of protease by active cancer ‘cells’ |
| kOPEN | 5 x 10-4 M-1 | Predicted overall rate coefficient for drug release – describing strand displacement process for opening of pore |
The modelling of the exchange process was further simplified by not considering the system in two/three dimensions. Avoiding the need to describe stochastic Brownian motion that would be particularly complex to predict in the ECM region. Therefore, once in the ECM, the only possible behaviour possible is diffusion in the one dimension considered, exchange into the capillary or interaction between liposomes/drug molecules and protease.
The PDEs required to model the system’s dynamics were formulated based on the general Convection-Diffusion Equation, with ‘reaction’ terms incorporated describing the production/destruction of the species as they interact within the system.
$$ \frac{\partial C}{\partial t} = \vec{\nabla} \cdot (\vec{\nabla} C) - \vec{\nabla} \cdot (\vec{v}C) + R \ \xrightarrow{Simplify} \ \frac{\partial C}{\partial t} = D \vec{\nabla}^2 C -\vec{v} \cdot \vec{\nabla} C + R $$The generalised form can then be simplified for our system, through the assumption that blood is incompressible, removing the divergence of velocity term. For blood flow in capillaries, it is not ideal to apply this assumption, however, as the system is only examined in one-dimension and the blood flow is assumed steady, the derivative of it will still be zero if incompressibility was not assumed. As previously noted, diffusivity of all species is constant in time and space, removing the gradient of diffusivity term. Reducing the equation above as described and formulating it for each specie forms the basis of PDEs to be solved.
Reducing the equation above as described and formulating it for each specie forms the basis of PDEs to be solved.
$$ \frac{\partial d_{ECM}}{\partial t} = D_{d-ECM} \nabla^2 (d_{ECM}) + R_1 \\ \frac{\partial d_{C}}{\partial t} = D_{d-C} \nabla^2 (d_{C}) - \vec{u} \cdot \vec{\nabla}{d_C} + R_1 \\ \frac{\partial L_{ECM}}{\partial t} = D_{L-ECM} \nabla^2 (L_{ECM}) + R_3 \\ \frac{\partial L_{C}}{\partial t} = D_{L-C} \nabla^2 (L_{C}) - \vec{u} \cdot \vec{\nabla}{L_C} + R_4 \\ \frac{\partial P}{\partial t} = D_{P} \nabla^2 (P) + R_5 $$Diffusion is described in these equations by the diffusivity multiplying the Laplacian of specie concentration, while the convection is the dot product of bulk velocity and gradient of specie concentration. The reaction terms, ‘R’, noted in these equations, were mathematically formulated based on the reaction pathways described in figure 2.
 $$
R_1 = \frac{d(d_{ECM})}{dt} = k_{d-Ex-ECM}(d_C) - k_{d-Ex-ECM} (d_{ECM}) + 50k_{OPEN}(L_{ECM})P - death_d (d_{ECM}) + death_L (L_{ECM}) \\
R_2 = \frac{d(d_{C})}{dt} = k_{d-Ex-C}(d_{ECM}) - k_{d-Ex-ECM} (d_C) - death_d (d_C) + death_L (L_C) \\
R_3 = \frac{d(L_{ECM})}{dt} = k_{L-Ex-C}(L_C) - k_{L-Ex-C} (L_{ECM}) + k_{OPEN}(L_{ECM})P - death_L (L_{ECM}) \\
R_4 = \frac{d(L_{C})}{dt} = k_{L-Ex-C}(L_{ECM}) - k_{L-Ex-ECM} (L_C) - death_L (L_C) \\
R_5 = \frac{dP}{dt} = \{Tumour state \} k_{P-Prod} - death_P (P)
$$
$$
R_1 = \frac{d(d_{ECM})}{dt} = k_{d-Ex-ECM}(d_C) - k_{d-Ex-ECM} (d_{ECM}) + 50k_{OPEN}(L_{ECM})P - death_d (d_{ECM}) + death_L (L_{ECM}) \\
R_2 = \frac{d(d_{C})}{dt} = k_{d-Ex-C}(d_{ECM}) - k_{d-Ex-ECM} (d_C) - death_d (d_C) + death_L (L_C) \\
R_3 = \frac{d(L_{ECM})}{dt} = k_{L-Ex-C}(L_C) - k_{L-Ex-C} (L_{ECM}) + k_{OPEN}(L_{ECM})P - death_L (L_{ECM}) \\
R_4 = \frac{d(L_{C})}{dt} = k_{L-Ex-C}(L_{ECM}) - k_{L-Ex-ECM} (L_C) - death_L (L_C) \\
R_5 = \frac{dP}{dt} = \{Tumour state \} k_{P-Prod} - death_P (P)
$$
The first reaction term accounts for each liposome containing multiple drug molecules through multiplying the second order term by 50. The coefficient of 50 is based upon calculating the internal volume of a liposome with 150 nm diameter and finding the ratio of this with the volume of a Cisplatin molecule with diameter 30 nm. As the ratio found doesn’t account for the molecular forces between Cisplatin molecules, leaving empty space within the liposome, and assumes that a liposome is filled to maximum capacity when formed, the outputted value of ~100 was reduced to reasonably account for these factors.
Therefore, using the two sets of equations developed, the dynamics of the system can be described. These can be solved through applying initial conditions for the concentrations of the species and reasonable boundary conditions to describe the physical system, producing the evolution of the five species’ concentration over a set time period.
With the system of equations established, solving them required initial and boundary conditions to be defined.
| Specie | Initial Condition | Relevance |
|---|---|---|
| dECM | [ dECM ] = 0 for all space | No liposomes have entered the system that could have released the drug |
| dC | [ dC ] = 0 for all space | " |
| LECM | [ LECM ] = 0 for all space | No liposomes have exchanged from capillary |
| LC | [ LC ] = 0 for all space except for spatial point two | Dose of drug carrying liposomes enters system at time zero – point two used due to limitation of model when applied to entrance of system |
| P | [P] = 0 for all space outside of active tumour region | Simulating a tumour being present that is already metastasizing surrounding tissue |
| Boundary Condition | Relevance |
|---|---|
| First spatial derivatives = 0 for all species at entrance of system | Neumann boundary condition represents that material flux entering system is zero – as the arrival of liposomes is described within the system as an initial condition |
| First spatial derivatives not restricted for all species at end of system | This enables the flux of material out of the system as expected through a non-prescribed first derivative value |
| Second spatial derivatives at end boundary equal to derivative at previous point | Avoids unreasonably restricting concavity of concentration profiles as calculation of it requires concentration at the x, x + δx and x – δx points – the x + δx point does not exist at end boundary |
| Initial third of spatial points have no exchange between regions | Allows distribution of vesicles to spread due to flow in capillary – better representing the concentration profile that would arrive into in vivo capillaries |
With these conditions the solver can now evaluate the concentration of each specie across the domain of each instance of time considered, outputting the desired dynamics. Referring to the system of PDEs, both the first and second order spatial derivatives need to be calculated, as well as calculating the reaction terms.
For the solver developed in this project, a forward-difference method of calculating first order derivatives was not compatible. However, Newton’s difference quotient can still be used by utilising a backward-difference approach, as shown below, because this produces identical derivative calculations when the step size is suitably small and was compatible with our solver.
$$ f'(x) = \lim_{\delta x \rightarrow 0} \left\{ \frac{f (x + \delta x)-f(x)}{\delta x}\right\} \\ f''(x) = \lim_{\delta x \rightarrow 0} \left\{ \frac{f (x + \delta x) + f(x - \delta x) -2f(x)}{{\delta x}^2}\right\} $$With the second order derivatives calculated in a similar way, avoiding limitations of the numerical solving approach through the appropriate boundary conditions when points \(x + \delta x\) and \( x - \delta x \) do not exist. The functions in these definitions represent the concentration of the specie being examined at the specific spatial point. Therefore, these are the values of the concentration at that specific time instance, known through calculating the changes in their value at each time point evaluated, starting at the initial values provided.
The reaction terms can simply be calculated using the concentration values known for that time instance. With these steps completed, the change in concentration with respect to time can be calculated by inputting the calculated values for the unknowns into the PDE (previously described) for each specie. Concentrations are then known for the t+n (where n is the constant time step size) time instance through the time derivative values calculated at the previous time instance and this process can be iteratively completed for the time period specified.
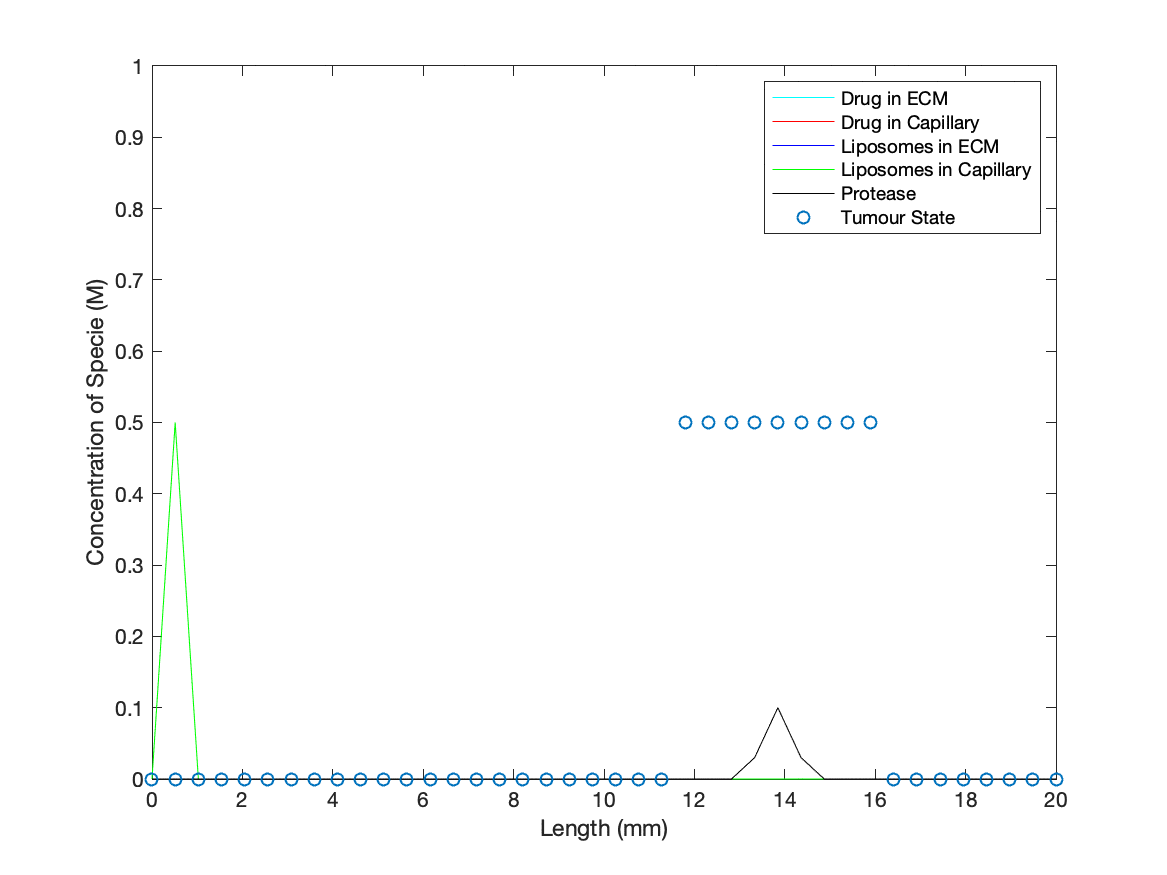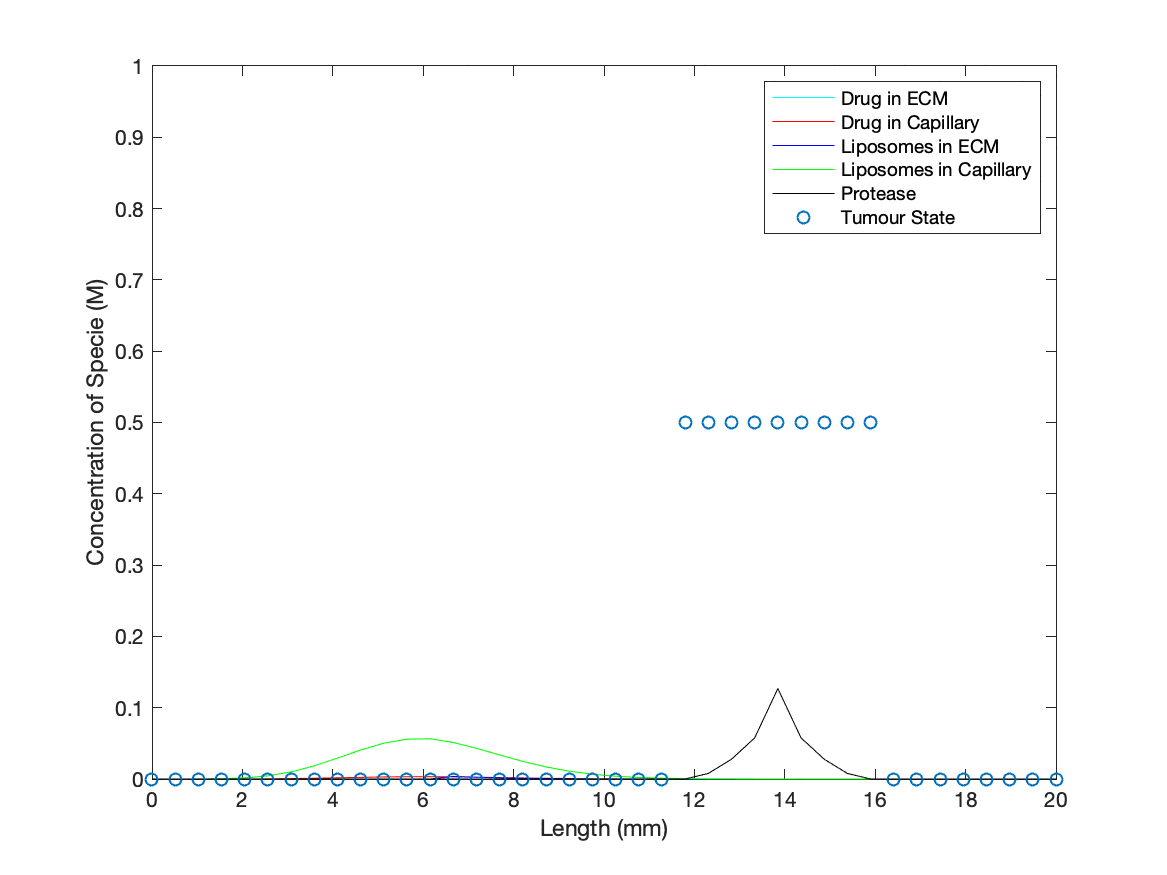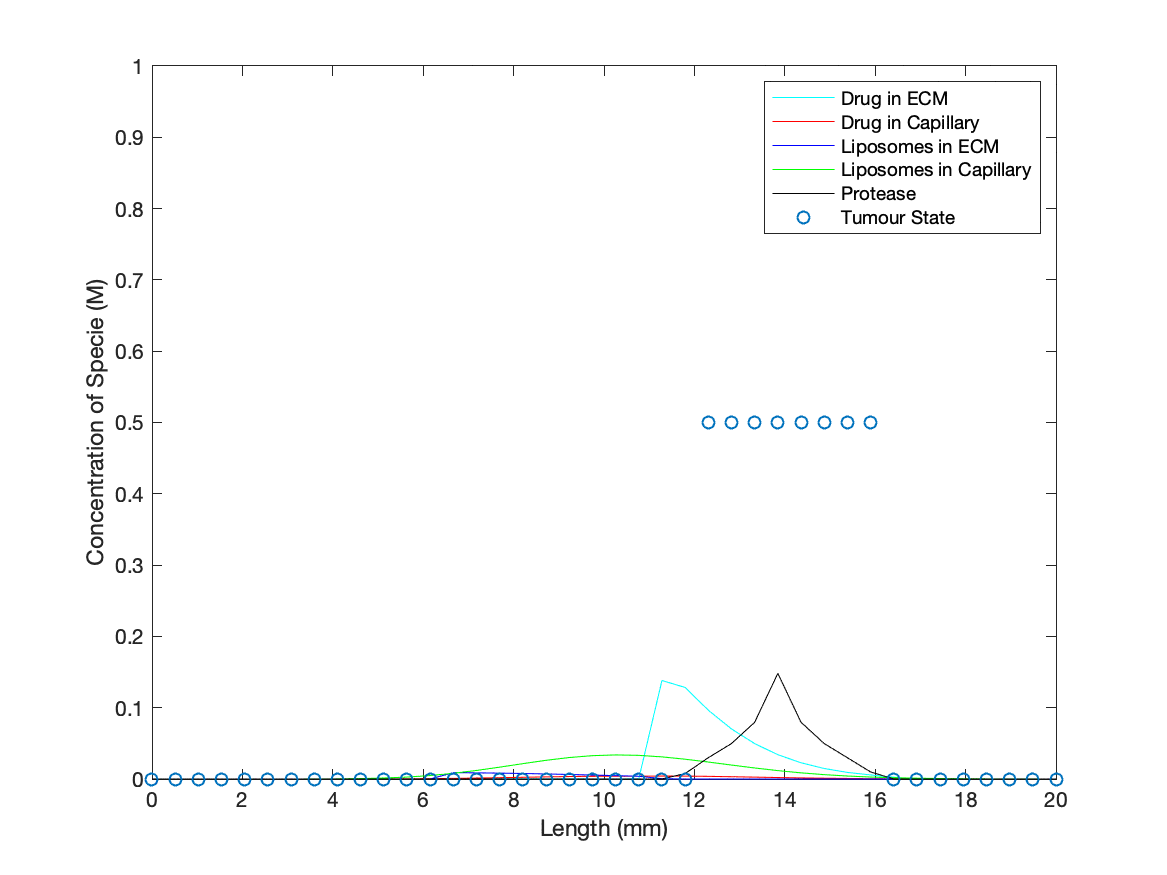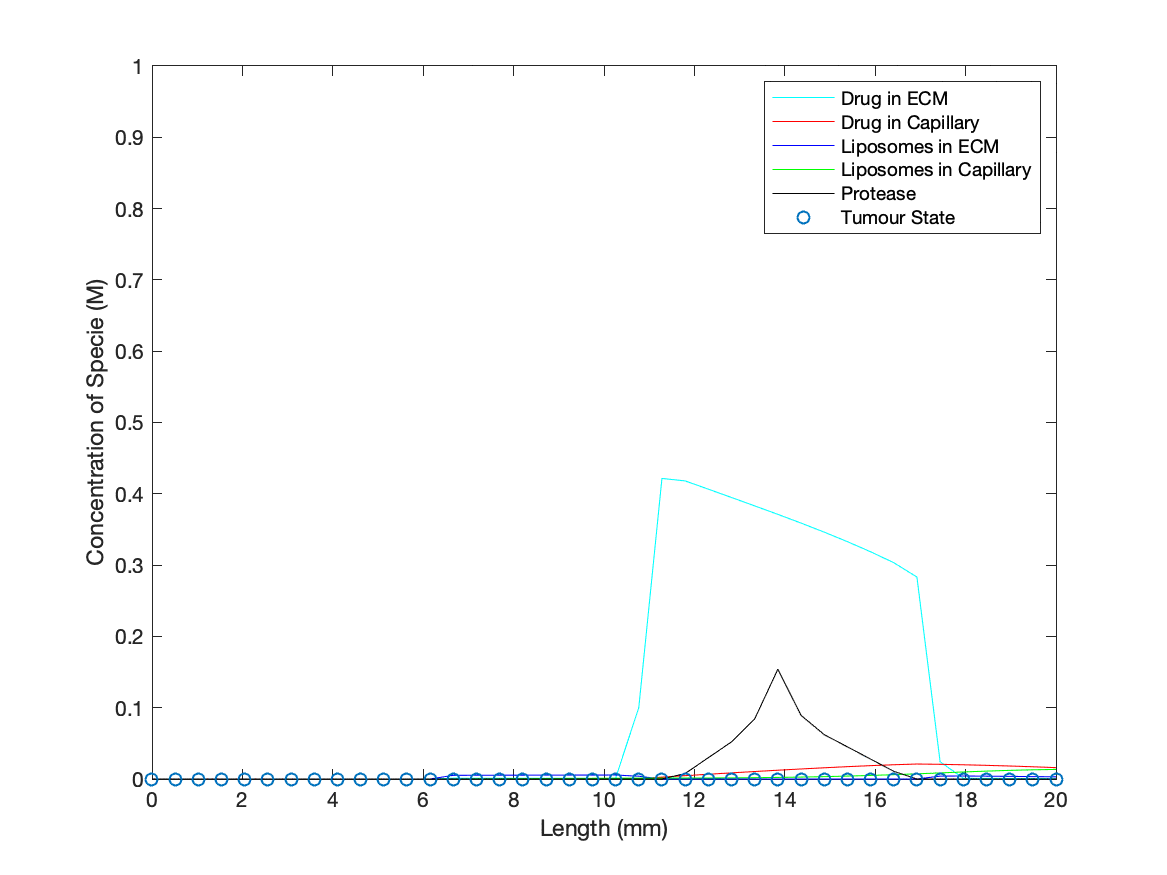
To add a dynamic cancer presence in the model, a ‘binary’ tumour was introduced into the ECM that was initialised as active at specified spatial points for t = 0. Protease concentration at points with an active tumour state will increase at a constant rate, simulating the production of the protease (biomarkers for our drug delivery method) by cancer cells. As the protease concentration rises, it may reach the threshold value to activate an adjacent point/’cell’ into switching to an active tumour state/’cancer cell’, producing an idealised cancer growth description. The values designated for production rate and threshold for spreading were based on giving quantitative ‘cancer’ growth during the run time of the simulation to demonstrate the dynamics of in vivo interactions between our technology and a cancer.
Cancerous cell death in the presence of the medicinal drug was represented by the tumour state being deactivated, determined through evaluating drug concentration against a threshold for medicinal activity across every point in space. The rate at which the drug is released, and subsequently 'kills' the cancer, is based upon the rate coefficient found from modelling the TMSD reactions involved in opening the nanopore.
Despite being modelled as a 3-step process, it has been shown (Mills
& Yurke 2003 ) that toehold-mediated strand displacement can be
approximated by bimolecular reactions with second-order rate
constants. This can be displayed as: $$ X(m,n) + S \quad
\overset{k_{(\beta^m, \beta_m, \gamma^n)}}{ \underset{k_{(\gamma^n,
\beta_m, \beta^m}}{\rightleftharpoons}} \quad Y + L(m,n) $$ where
these rate constants are related to the constants defined previously
by: $$k_{(\beta^m, \beta_m, \gamma^n)} \equiv \frac{k_{r(\beta^m)}
k_f k_b}{k_{r(\gamma^n)}k_{r(\beta^m)} + k_{r(\gamma^n)}k_b +
k_{r(\gamma^m)}k_b} $$ To ensure that these simulations are accurate
to within 50%, the concentration of \( X(m,n) \) must be kept below
a certain critical concentration which for our purposes is \( 100nM
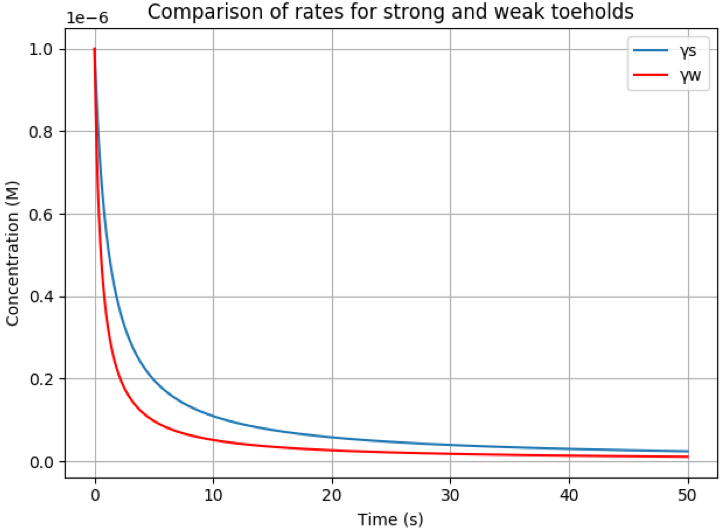
- 1\mu M \). Below is a simulation of this process with the rate
constants calculated for the two different toeholds:
The rate of this reaction also strongly depends on the composition
of the toehold. Most estimates are made using mixed teoholds but
toeholds can also be assigned as strong (exclusively G/C pairs) and
weak (exclusively A/T). This results in a quantifiable difference in
binding energy and therefore reaction rates due to the difference in
number of Hydrogen bonds.
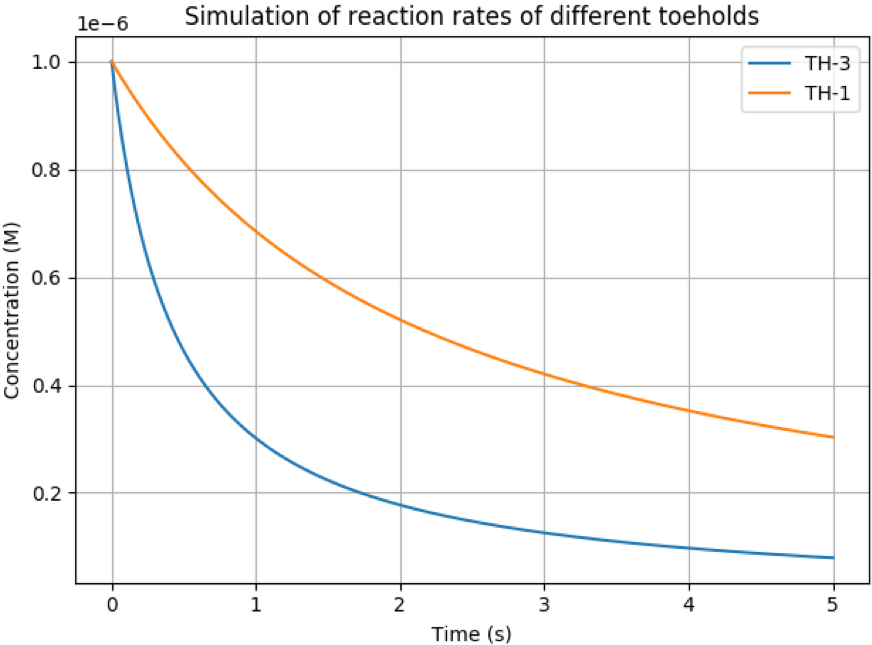
For the purposes of illustrating this difference, below is plotted
the reaction rates for two domains of length m=6 with one composed
of only G/C pairs and the other only with A/T pairs. It is clear to
see that after 3 seconds there is a distinct difference in the rate
at which this reaction occurs.